Respiratory syncytial virus sequesters NF-κB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signalling
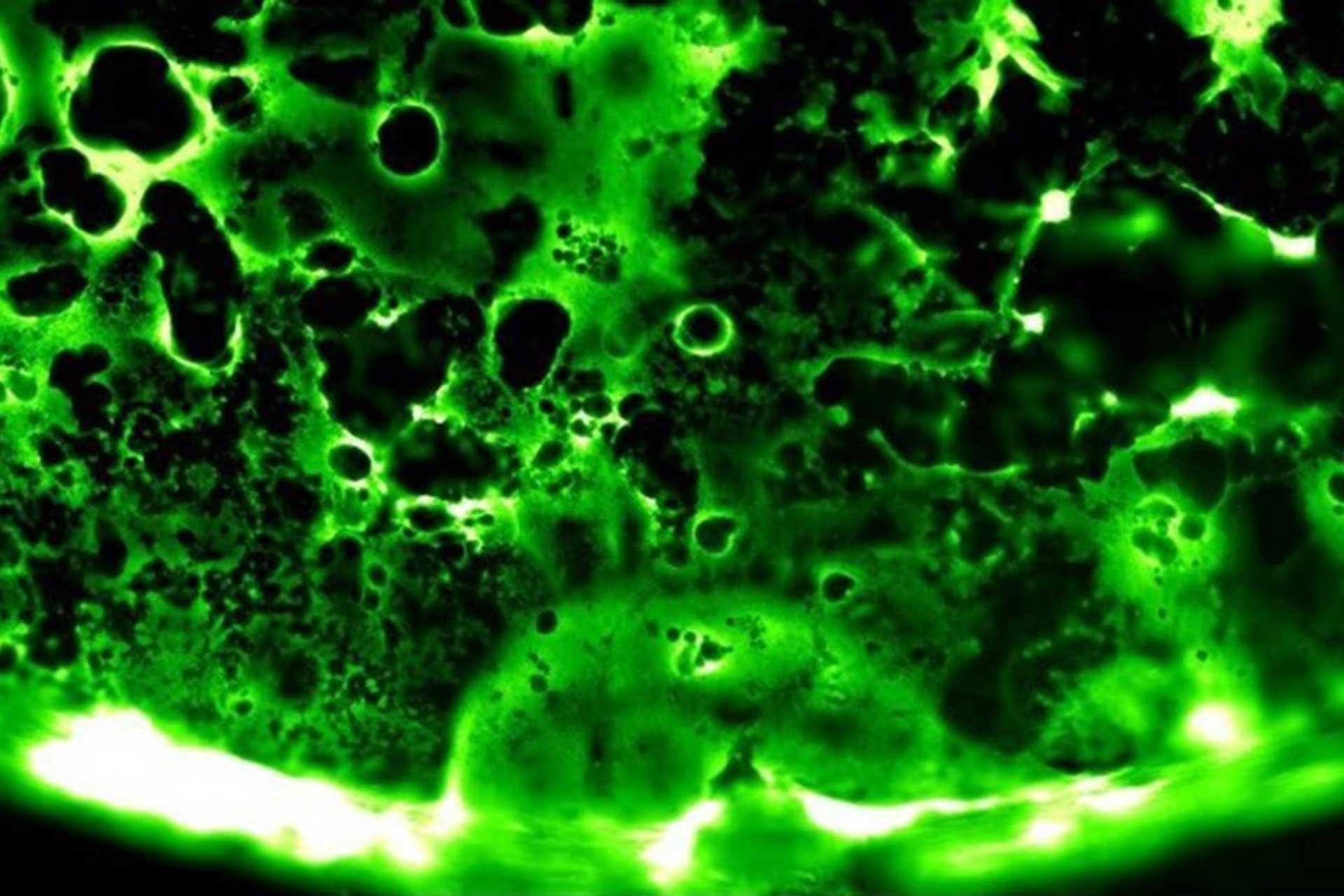
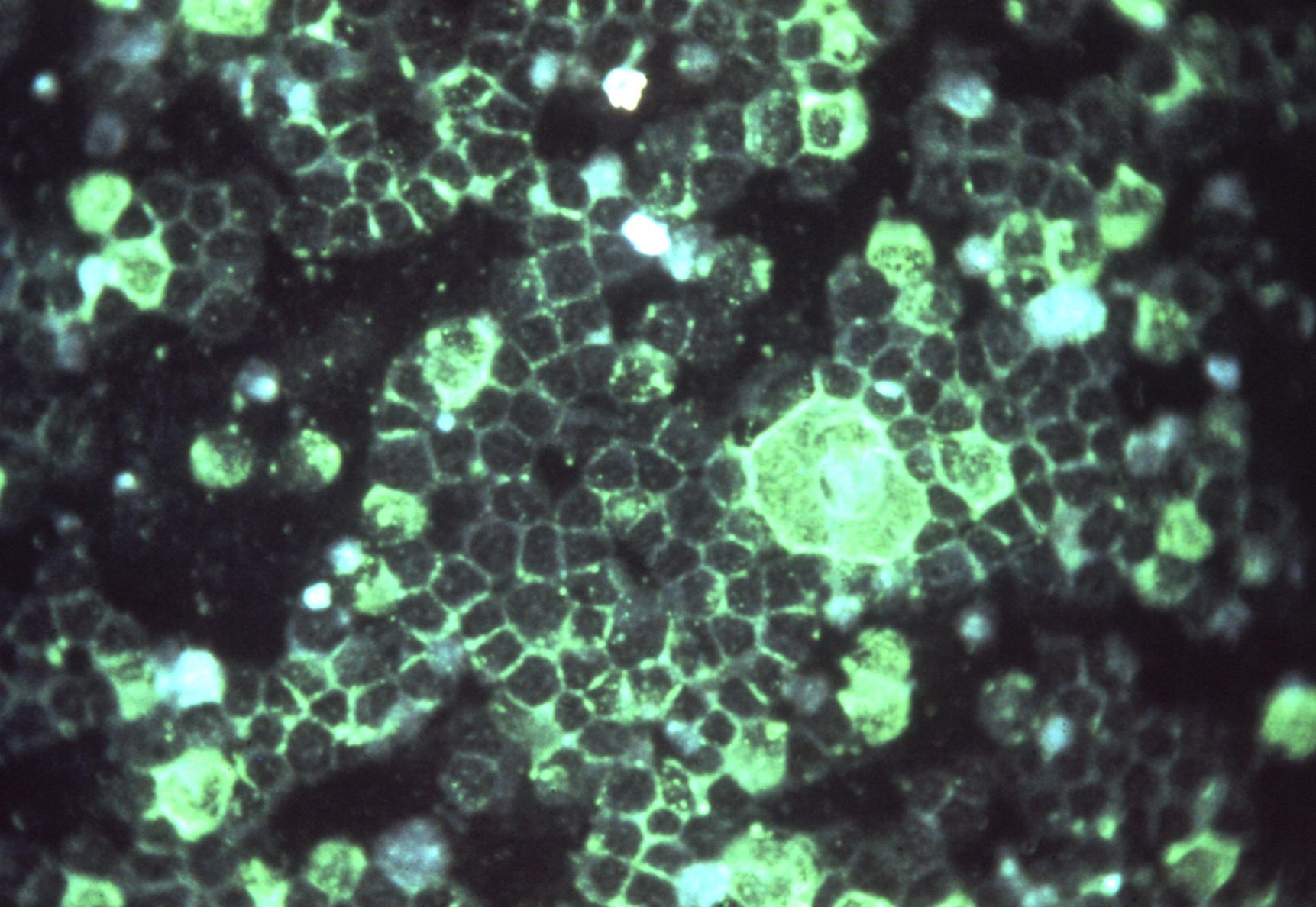
Viruses routinely employ strategies to prevent the activation of innate immune signalling in infected cells. RSV is no exception, encoding two accessory proteins (NS1 and NS2) which are well established to block Interferon signalling. However, RSV-encoded mechanisms for inhibiting NF-κB signalling are less well characterised. In this study we identified RSV-mediated antagonism of this pathway, independent of the NS1 and NS2 proteins, and indeed distinct from other known viral mechanisms of NF-κB inhibition. In both human and bovine RSV infected cells we demonstrated that the P65 subunit of NF-κB is rerouted to perinuclear puncta in the cytoplasm, puncta which are synonymous with viral inclusion bodies (IBs), the site for viral RNA replication. Captured P65 was unable to translocate to the nucleus or transactivate a NF-κB reporter following TNF-α stimulation, confirming the immune-antagonistic nature of this sequestration. Subsequently, we used correlative light electron microscopy (CLEM) to colocalise RSV N protein and P65 within bRSV IBs; granular, membraneless regions of cytoplasm with liquid organelle-like properties. Additional characterisation of bRSV IBs indicated that although they are likely formed by liquid-liquid phase separation (LLPS), they have a differential sensitivity to hypotonic shock proportional to their size. Together, these data identify a novel mechanism for viral antagonism of innate immune signalling which relies on sequestration of the NF-κB subunit p65 to a biomolecular condensate – a mechanism conserved across the Orthopneumovirus genus and not host-cell specific. More generally they provide additional evidence that RNA virus IBs are important immunomodulatory complexes within infected cells.
Importance: Many viruses replicate almost entirely in the cytoplasm of infected cells; however, how these pathogens are able to compartmentalise their life cycle to provide favourable conditions for replication and to avoid the litany of antiviral detection mechanisms in the cytoplasm remains relatively uncharacterised. In this paper we show that bovine RSV (bRSV), which infects cattle, does this by generating inclusion bodies in the cytoplasm of infected cells. We confirm that both bRSV and human RSV viral RNA replication takes place in these inclusion bodies, likely meaning these organelles are a functionally conserved feature of this group of viruses (the orthopneumoviruses). Importantly, we also showed that these organelles are able to capture important innate immune transcription factors (in this case NF-KB), blocking the normal signalling processes that tell the nucleus the cell is infected, which may help us to understand how these viruses cause disease.