Our group
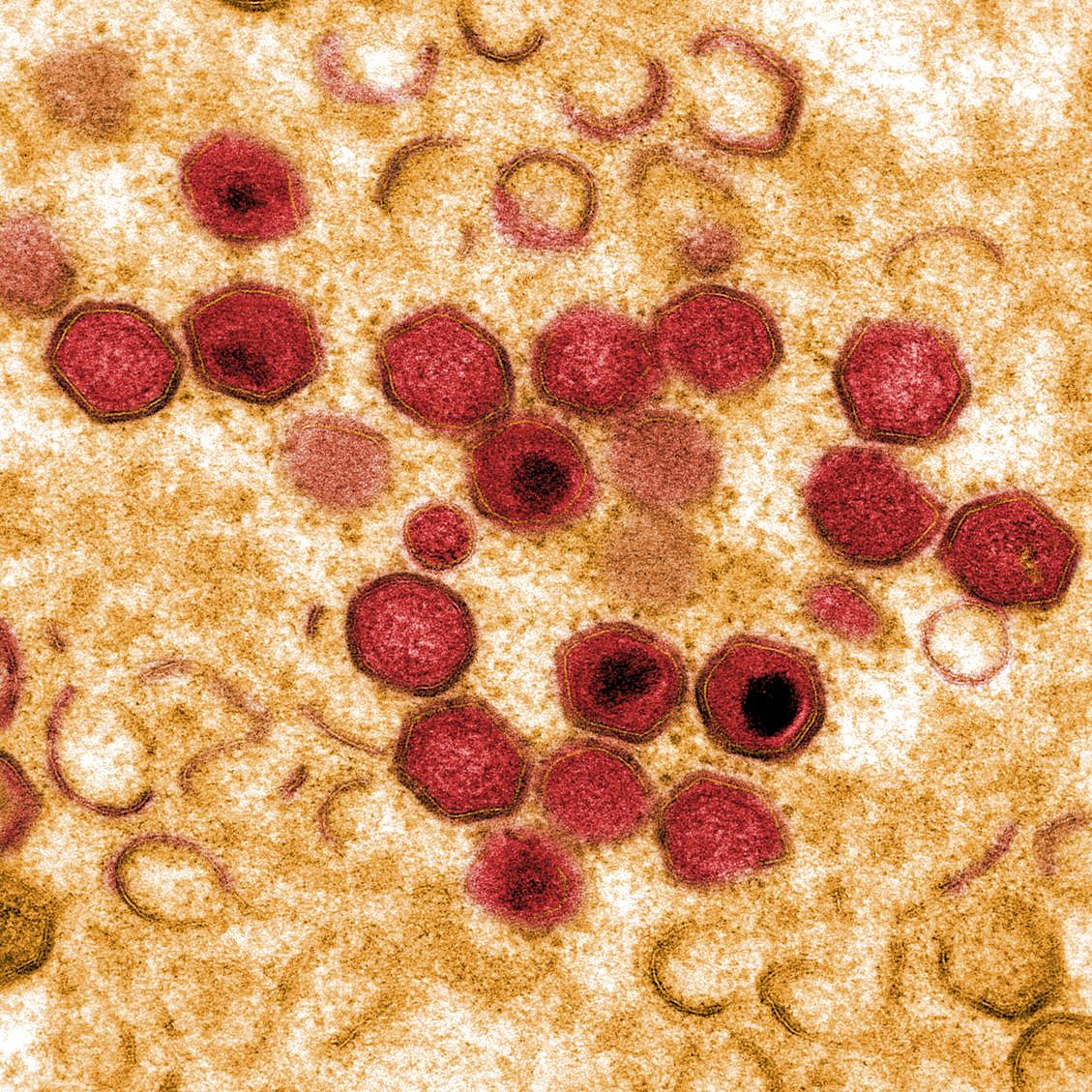
African swine fever virus (ASFV) causes a haemorrhagic fever in domestic pigs and wild boar resulting in mortality rates up to to 100%. In contrast, hosts in the ancient sylvatic cycle in E Africa, warthogs, bushpigs and soft ticks of Ornithodoros spp can be persistently infected with few clinical signs.
ASF is endemic in sub-Saharan Africa and Sardinia, and since 2007 has spread through the Trans-Caucasus, Russian Federation and into Eastern Europe.
The disease has a huge impact socially and economically, posing a threat to global food security. The lack of a vaccine limits options for disease control.
ASFV is a large predominantly cytoplasmic DNA virus which encodes 150 to 167 open reading frames. These include a large number that are not essential for virus replication but encode proteins that have important roles in host interactions including in immune evasion.
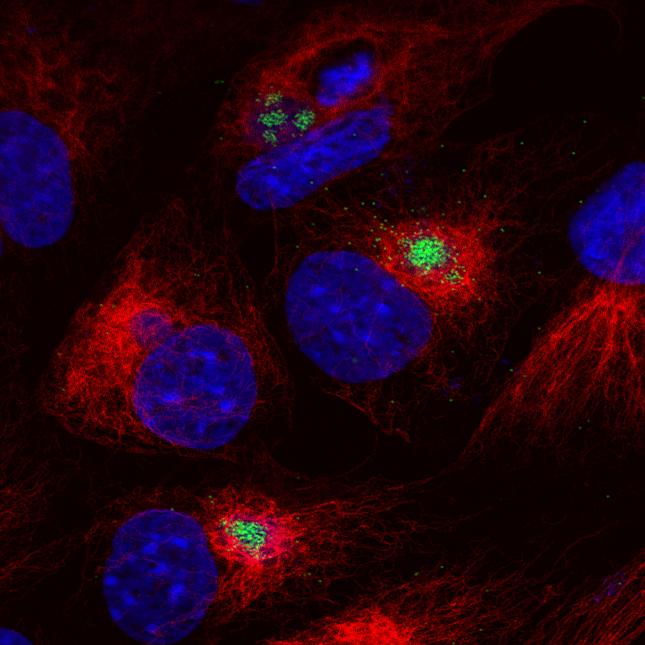
The virus replicates in macrophages, key cells involved in activating innate and adaptive immune responses. Modulation of macrophage function by ASFV is critical both for mechanisms of pathogenesis and immune evasion.
Our aims
The aim of our research is to improve understanding of ASFV host interactions at the molecular, cellular and whole animal level to underpin work leading to improved disease control, including development of vaccines.
The objectives are to:
- Use a functional genomic approach to characterise ASFV proteins involved in immune evasion and virulence and define the molecular targets of the proteins
- Investigate the role of ASFV proteins on modulating host responses during infections of cells and whole animals
- Apply this knowledge to develop candidate vaccines through targeted gene deletions
- Identify protective antigens (in collaboaration with the Vaccinology Group)
Our research
- Investigation of the mechanisms by which ASFV inhibits induction of type I interferon (IFN) antiviral host responses.
The ASFV molecular patterns (PAMPS) and the host pattern recognition receptors that can lead to activation of type I IFN.are under investigation.
In addition, several ASFV proteins that inhibit IFN induction have been identified. Using gene deleted ASFV the impact of these proteins on host responses during in vitro infection of cells and on virus pathogenesis and induction of protective immunity in pigs is being studied.
- Characterisation of ASFV proteins that inhibit host defences.
By functional screening, using a plasmid library, novel inhibitors of stress-activated pathways have been identified and their mode of action investigated by identifying interacting host proteins.
A similar approach is being used to identify inhibitors of other host pathways including the NFkB pathway.
- ASFV genomics and transcriptomics.
Three complete ASFV genome sequences have been determined to identify differences between virulent and attenuated strains and characterise the strain introduced into Georgia in 2007.
Procedures are being established for high throughput next generation sequencing of genomes and RNA seq to analyse transcriptional responses following infection.
- Evaluation of ASFV candidate vaccine strains.
ASFV strains, attenuated by gene deletions or naturally attenuated are being evaluated by immunisation of pigs and lethal challenge. The routes and doses of immunisation are being compared and correlates of protection and pathogenesis studied.
The long term persistence of virus and duration of immunity are also being evaluated. In addition cell lines suitable growth of virus are under evaluation.
- Dynamics of ASFV transmission.
The dynamics of direct and indirect transmission of ASFV were studied as to provide data for modelling the spread of disease within and between pens on infected farms.
Timing and survival of virus in excretions from infected pigs were obtained to provide data for risk analysis.
Our impact
African swine fever virus causes an acutely fatal haemorrhagic fever in pigs which has a high socio-economic impact. The lack of a vaccine limits options for disease control.
The group has contributed to knowledge on genes encoded by ASFV by complete genome analysis of high and low virulence isolates and the strain currently circulating in the Russian Federation and Eastern Europe. This has provided information to identify genes involved in virulence, immune evasion and virus replication.
The knowledge has been applied to development of candidate vaccines through rational gene deletions to produce candidate live attenuated vaccines and by expression of proteins to identify protective antigens, working with GALVmed to identify companies to work with and ensure any vaccines produced will be globally accessible.
As national and WOAH disease expert, Linda Dixon provides advice to the UK and other countries.