Our group
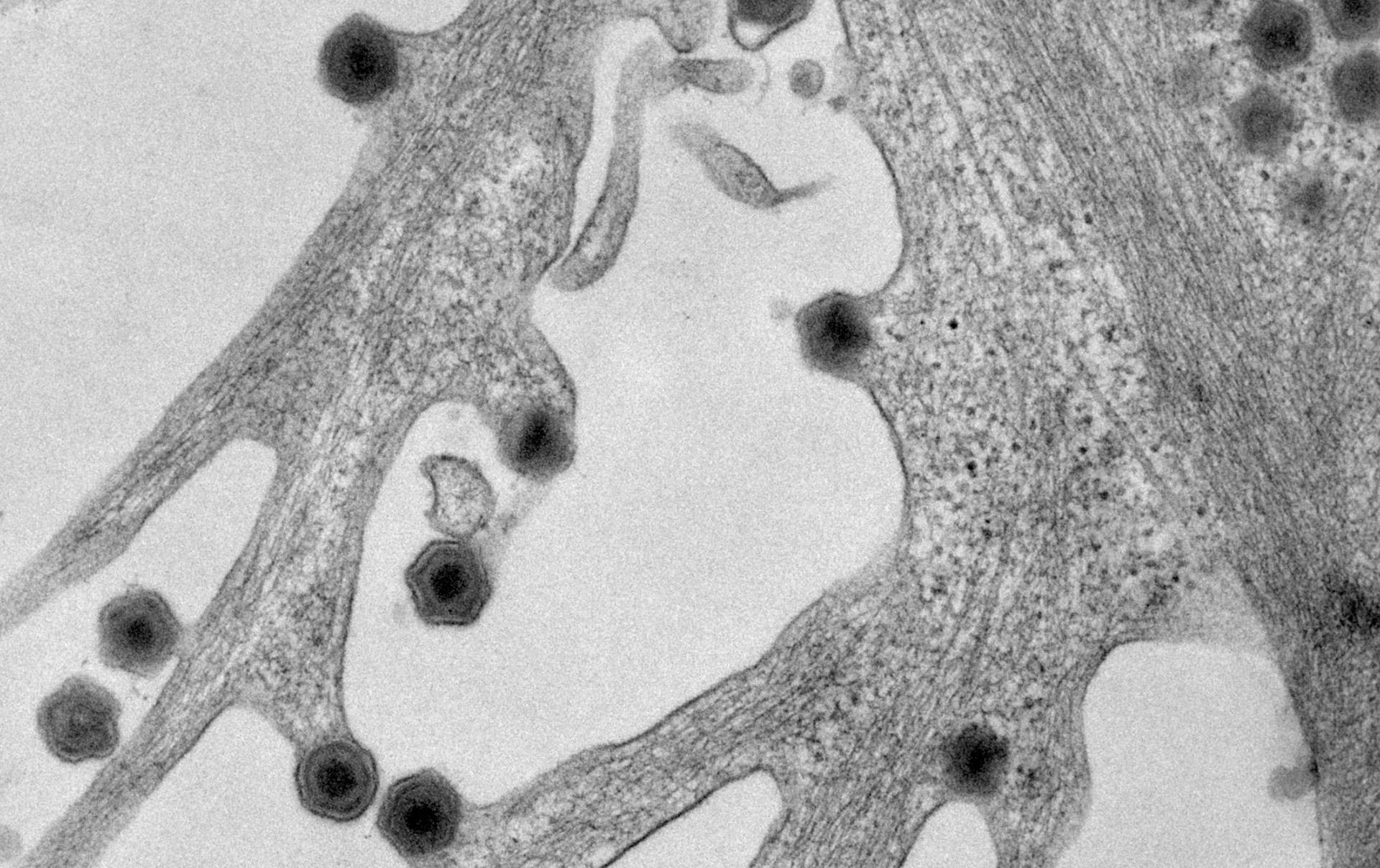
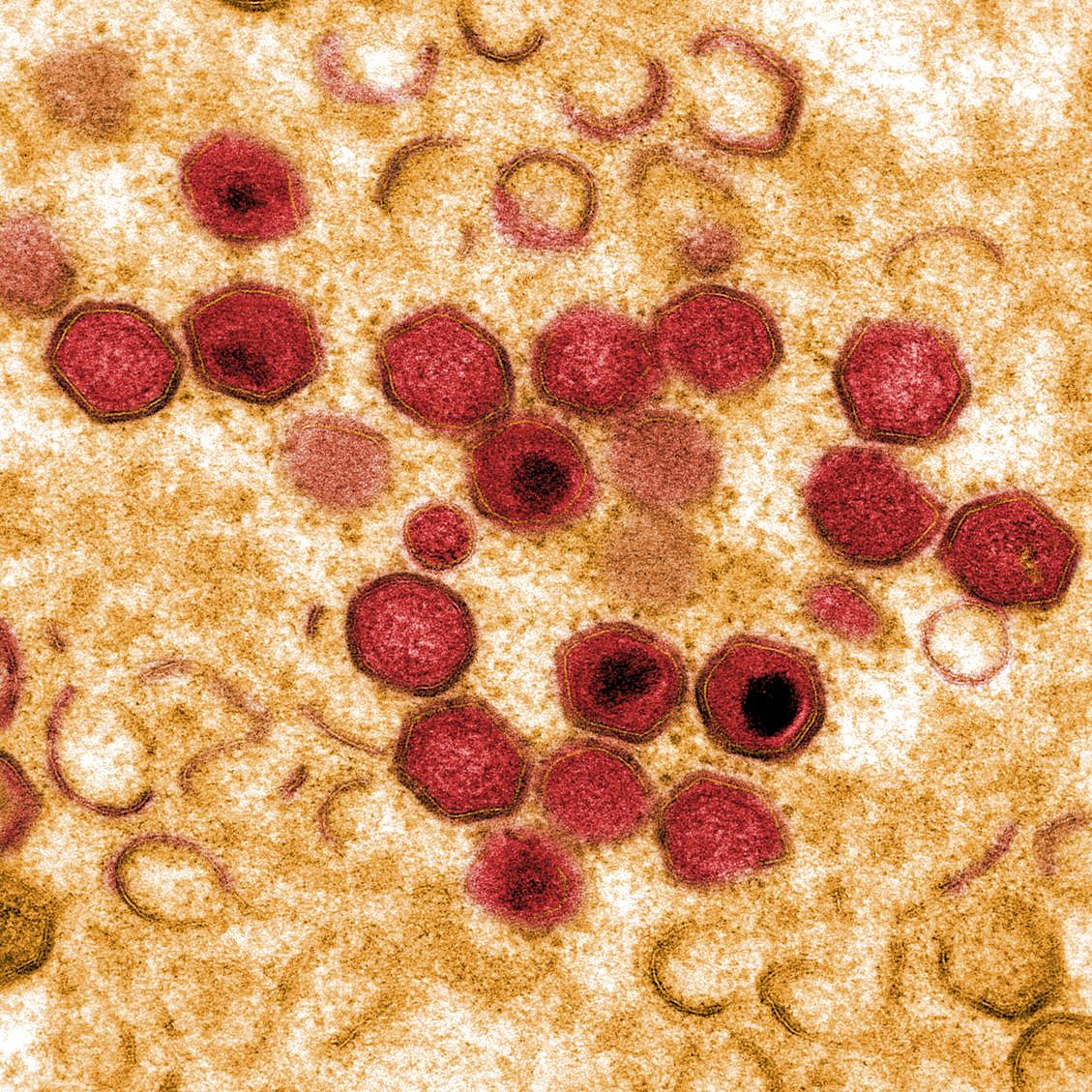
African Swine Fever Virus (ASFV) is a high consequence pathogen, with a specific tropism for cells from the monocyte/macrophage lineage, which are key for both the host innate and adaptive immune responses. The virus manipulates these responses for its own benefit and around 1/3 of ASFV encoded genes are predicted to have evolved to modify key immunoregulatory pathways. The “ASFV Immune Evasion group” focuses on characterising virus-host interactions at molecular level in both infected and bystander cells and understanding the consequences of these interactions for viral attenuation and disease pathogenesis.
Our aims
The aim of our group is to provide fundamental knowledge on ASFV molecular mechanisms of virulence, by dissecting the complexity of its interactions with the host cells. This will underpin the development of ASF control strategies such as vaccines and antivirals. Additionally, identification of “molecular signatures” associated with different types of disease, including chronic ASF, will allow us to predict “phenotype from genotype”, assisting epidemiological surveillance in regions where naturally attenuated viruses might emerge.
Our research
- Understand the role of cooperation or competition between members of the multigene families (MGF) during viral infection.
Members of the MGF360 and MGF505 are inhibitors of type I Interferon (IFN) and other host innate responses, and known virulence factors. We are using a panel of ASFV gene deletion mutants with single and multiple deletions to explore the contribution of different MGFs for the inhibition of innate responses. We are characterising both the induction of Interferon Stimulated Genes (ISGs) and the virus replication in the presence of chemical inhibitors of relevant cellular pathways. We are also using a quantitative proteomics approach to identify differences in host proteins between cells infected with the wild-type virus and the deletion mutants.
- Characterise the role of cell-cell interactions in the modulation of host immune responses.
ASFV encodes proteins that are expressed at the cell surface, providing the opportunity for the virus to manipulate the function of bystander cells. Some of these proteins were already described but their mechanisms of action remain poorly understood. We will use a plasma membrane proteomics approach to identify other viral proteins belonging to this category. We are also developing tools and reagents to further characterise their function with a special emphasis on bystander immune cells.
- We are part of an international consortium that uses high throughput approaches to identify new interactions between ASFV and host proteins. We are further characterising these interactions in the context of viral infection, by producing deletion mutant viruses and evaluating their replication and impact on cellular signalling cascades.
Our impact
African swine fever (ASF) is a transboundary disease of domestic pigs characterised by a severe haemorrhagic syndrome. Lethality can reach 100% in susceptible pig populations, resulting in devastating economic losses. ASF is endemic in most countries of sub-Saharan Africa and in Sardinia. In 2007, ASF was introduced to the Caucasus region and spread towards Central-eastern Europe and Asia, recently reaching the American continent thus becoming a global challenge.
Effective vaccines and antivirals against ASF would have an enormous impact on affected countries, securing the livelihoods of those involved in the pig industry. They would also contribute to prevent further global spread of the virus, therefore protecting the UK from future disease incursions. This can only be achieved by a deeper understanding on how ASFV evades the host immune responses, providing targets for genetic manipulation and/or drug development.